SiteMapper
Explore Binding Sites
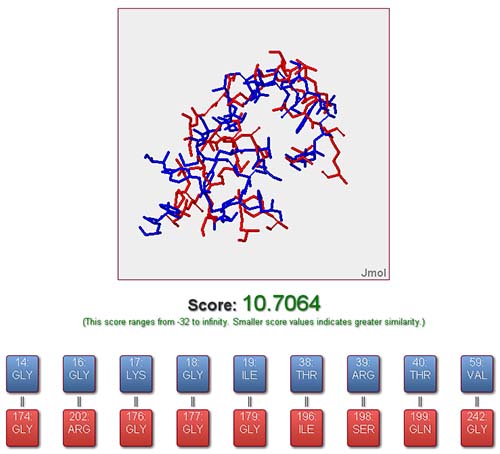
SiteMapper is a free webserver originally designed for computational evaluation of the binding site similarity of proteins and analysis of the residue correspondences based on 3Dstructure information. By means of structure-based alignment, SiteMapper is capable of detecting common patterns or residue reservation between binding sites which are not required to possess continuous residues or to be homologous. These common patterns can be further used to perform protein classification or reverse virtual screening, e.g.:
Internally, the SiteMapper is implemented based on an in-house developed program which abstracts the input binding sites as backbones of alpha carbons and tries to align these 3D structures. The underlying alignment process is deviation induced and an efficient down-hill simplex algorithm is applied to search the optimal alignment which takes both the residue position and residue type into full consideration, while the residue correspondence is identified using a Hungarian algorithm based method. SiteMapper can be very useful for evaluating the binding site similarity or performing discontinuous sequence alignment for binding sites in a 3D manner.
In order to evaluate a pair of binding sites, user should provide a valid PDB code and identifiers of residues representing the binding sites for both reference and fit structures. The input interface includes the following fields to define a binding site:
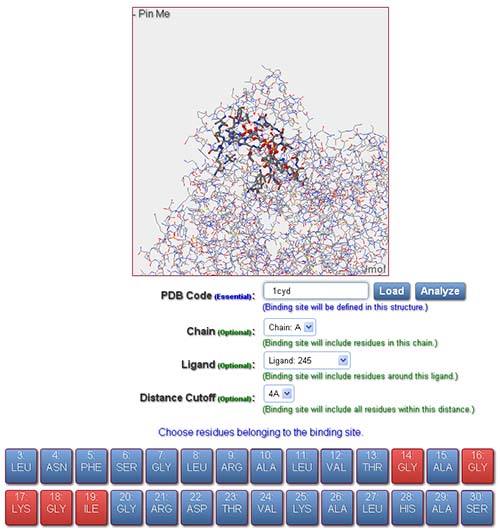
In order to define a binding site for subsequent similarity evaluation, the following operations should be performed:
Once both the reference and fit binding site have been defined, it is ready to submit the job to evaluate the similarity. Click the “Submit” button to submit the job and the user will be automatically directed to a job status page which provides detailed information about a job. Of all the information displayed in the job status page, the most important one is the job ID which should be reserved by the user in order to query the status or get the result of a job.

Note: If user provides an email address at the submission of a job, then a notification email will be sent to the corresponding address once the job is completed.
Through the “Result” item in the navigation bar located at the top of main page, user can query the status of a job conveniently and view the result if the job is completed. Provide the job ID and corresponding email address if any, user will be directed to the job status page. The job status page displays detailed information about a job, e.g. current status of the jobs, links to view or download result. There are several possible statuses for a job, as follows:

Note: The job status page will automatically refresh itself to reflect the most recent status of the job.
From the job status page, the result of a job could be viewed once it is completed. The online output provided by SiteMapper is a visualization of the aligned binding sites along with the similarity score and the residue correspondences between the reference and query binding sites. Each residue correspondence could be highlighted and visualized in 3D mode freely by the user. Additionally, all result files (including files for both original and aligned structure, files containing residue correspondences and other information) are packaged and ready to be downloaded by user for performing further analysis locally.