


PharmMapper Server: An updated integrated pharmacophore matching platform with statistial method for potential target identification.
Drug target identification, which includes many distinct algorithms for finding genes and proteins, is the first step in drug discovery. When 3D structures of the targets are available, the problem of target identification is usually converted to finding the best interaction mode between the potential target candidates and probe small molecules. Pharmacophore, which is the spatial arrangement of features that is essential for a molecule to interact with a specific target receptor, is an alternative method despite of molecular docking for achieving this goal.
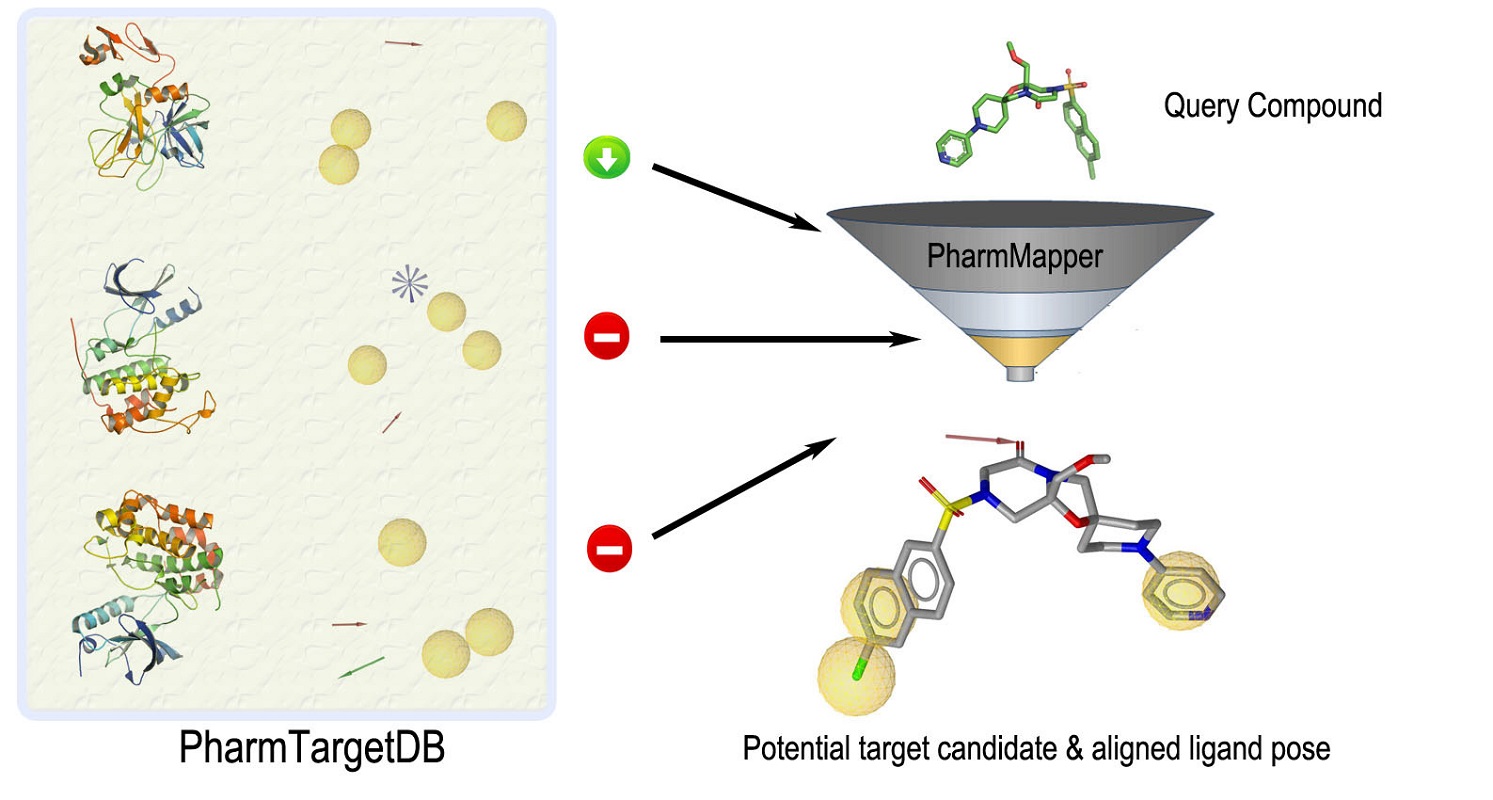
PharmMapper Server is a freely accessed web-server designed to identify potential target candidates for the given probe small molecules (drugs, natural products, or other newly discovered compounds with binding targets unidentified) using pharmacophore mapping approach. Benefited from the highly efficient and robust mapping method, PharmMapper bears high throughput ability and can identify the potential target candidates from the database within a few hours.
PharmMapper is backed up by a large, in-house repertoire of pharmacophore database extracted from all the targets in TargetBank, DrugBank, BindingDB and PDTD. Over 7,000 receptor-based pharmacophore models (covering 1,627 drug targets information, 459 of which are human protein targets) are stored and accessed by PharmMapper. PharmMapper finds the best mapping poses of the user uploaded molecules (in Tripos Mol2 or MDL SDF format) against all the targets in PharmTargetDB and top N potential drug targets as well as respective molecule's aligned poses are outputted.
In addition, to improve PharmMapper's accuracy further, we extracted all the pharmacophore model's ligands from PDB and stored them as a library ligand dataset in mol2 format and calculate these ligands' PharmMapper fit score. This process generates a nearly 7,000×7,000 score matrix. After this, when a new molecule is submitted, the fit score to each pharmacophore will be calculated first, and then every fit score of a specific pharmacophore will be compared to the fit score matrix to measure its score level among all the scores of the pharmacophore. This process adds up to the pure fit score with more statistical meaning and confidence comparing with random pharmacophore matching.
To use PharmMapper server, please upload your molecule file in Submit Job page. The job information as well as the structures of submitted molecules is invisible between different users. Here is a sample output of identified targets for Tamoxifen.
Current release. The current release, i.e. Version 2017, is based on the contents of PDB officially released by Jan 1st, 2016. This release applies Cavity1.1 to detect the binding sites on the surface of a given protein structure and rank them according to the corresponding druggability scores. A receptor-based pharmacophore modeling program Pocket 4.0 was then used to extract pharmacophore features within cavities. In this approach, a total of 23236 proteins covering 16159 pharmacophore models which are predicted as druggable binding sites and 52431 pharmacophore models with a pKd value higher than 6.0 are picked out, which is currently the largest collection of this kind. Compared to the last release (v.2010), target pharmacophore models included in this release have increased more than six times, from 7302 to over almost 53000.
Please notice that PharmMapper may NOT be used for commerical purposes.
Reference:
Xiaofeng Liu, Sisheng Ouyang, Biao Yu, Kai Huang, Yabo Liu, Jiayu Gong, Sisuan Zheng, Zhihua Li, Honglin Li, Hualiang Jiang. PharmMapper Server: a web server for potential drug target identification via pharmacophore mapping approach. Nucleic Acids Res., 2010, 38, W609-W614.
Xia Wang, Chenxu Pan, Jiayu Gong, Xiaofeng Liu, Honglin Li. Enhancing the Enrichment of Pharmacophore-Based Target Prediction for the Polypharmacological Profiles of Drugs. J. Chem. Inf. Model., 2016, 56, 1175-1183.
Xia Wang, Yihang Shen, Shiwei Wang, Shiliang Li, Weilin Zhang, Xiaofeng Liu, Luhua Lai, Jianfeng Pei, Honglin Li. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res., 2017, 45, W356-W360.